What is CrossMap ?
CrossMap is a program for genome coordinates conversion between different assemblies (such as hg18 (NCBI36) <=> hg19 (GRCh37)). It supports commonly used file formats including BAM, CRAM, SAM, Wiggle, BigWig, BED, GFF, GTF, MAF VCF, and gVCF.
How CrossMap works?

Release history
01/11/2024: Release version 0.7.0
Fix bugs for VCF varaints liftover.
Handle non-DNA ALT alleles such as <DEL>
Use pyproject.toml to replace “setup.py”.
Note
From v0.7.0 onwards, the main program CrossMap.py is now renamed to CrossMap
due to the restriction on using “.” in the script name in the “pyproject.toml” file.
To ensure compatibility with the previous pipelines, please include the following line in
your ~/.bashrc file.
alias CrossMap.py='CrossMap'
07/21/2023: Release version 0.6.4
Fix bug when the sequence in BAM file is represented as “*”
Change code style
07/12/2022: Release version 0.6.4
Fix bug when the input bigwig file does not have coverage signal for some chromosomes.
When the input VCF file does not have CONTIG field, use long chromosome ID (e.g., “chr1”) as default.
03/04/2022: Release version 0.6.3
Fix bug in v0.6.2. “Alternative allele is empty”
02/22/2022: Release version 0.6.2
For insertions and deletions, the first nucleotide of the ALT allele (the 5th field in VCF file) is updated to the nucleotide at POS of the reference genome
11/29/2021: Release version 0.6.1
Same as v0.6.0. Remove unused modules from the lib folder.
11/16/2021: Release version 0.6.0
Use
os.path.getmtimeinstead ofos.path.getctimeto check the timestamps of fasta file and its index file.Add ‘–unmap-file’ option to
CrossMap.py bedcommand.
4/16/2021: Release version 0.5.3/0.5.4
Add CrossMap.py viewchain to convert chain file into block-to-block, more readable format.
12/08/2020: Release version 0.5.2
Add ‘–no-comp-alleles’ flag to CrossMap.py vcf and CrossMap.py gvcf. If set, CrossMap does not check if the “reference allele” is different from the “alternative allele”.
08/19/2020: Release version 0.5.1
In CrossMap.py region: keep additional columns (columns after the 3rd column) of the original BED file after conversion.
08/14/2020: Release version 0.5.0
Add CrossMap.py region function to convert large genomic regions. Unlike the CrossMap.py bed function, which splits big genomic regions, CrossMap.py region tries to convert the big genomic region as a whole.
07/09/2020: Release version 0.4.3
Structural Variants VCF files often use INFO/END field to indicate the end of a deletion. v0.4.3 updates “END” coordinate in the INFO field.
05/04/2020: Release version 0.4.2
Support GVCF file conversion.
03/24/2020: Release version 0.4.1
Deal with consecutive TABs in the input MAF file.
10/09/2019: Release version 0.3.8
The University of California holds the copyrights in the UCSC chain files. As requested by UCSC, all UCSC-generated chain files will be permanently removed from this website and the CrossMap distributions.
07/22/2019: Release version 0.3.6
Support MAF (mutation annotation format).
Fix error “TypeError: AlignmentHeader does not support item assignment (use header.to_dict()” when lifting over BAM files. User does not need to downgrade pysam to 0.13.0 to lift over BAM files.
04/01/2019: Release version 0.3.4
Fix bugs when chromosome IDs (of the source genome) in chain file do not have ‘chr’ prefix (such as “GRCh37ToHg19.over.chain.gz”). This version also allows CrossMap to detect if a VCF mapping was inverted, and if so, reverse complements the alternative allele (Thanks to Andrew Yates). Improve wording.
01/07/2019: Release version 0.3.3
Version 0.3.3 is exactly the same as Version 0.3.2. The reason to release this version is that CrossMap-0.3.2.tar.gz was broken when uploading to pypi.
12/14/18: Release version 0.3.2
Fix the key error problem (e.g KeyError: “sequence ‘b’7_KI270803v1_alt’’ not present”). This error happens when a locus from the original assembly is mapped to an “alternative”, “unplaced” or “unlocalized” contig in the target assembly, and this “target contig” does not exist in your target_ref.fa. In version 0.3.2, such loci will be silently skipped and saved to the “.unmap” file.
11/05/18: Release version 0.3.0
v0.3.0 or newer will Support Python3. Previous versions support Python2.7.*
add pyBigWig as a dependency.
Installation
Install from PyPI
pip3 install CrossMap
Install from GitHub
pip3 install git+https://github.com/liguowang/CrossMap.git
Upgrade
pip3 install CrossMap --upgrade
Input and Output
Chain file
A chain file describes a pairwise alignment between two reference assemblies. UCSC and Ensembl chain files are available:
UCSC chain files
Chain files from hs1 (T2T-CHM13) to hg38/hg19/mm10/mm9 (ore vice versa): https://hgdownload.soe.ucsc.edu/goldenPath/hs1/liftOver/
Chain files from hg38 (GRCh38) to hg19 and all other organisms: http://hgdownload.soe.ucsc.edu/goldenPath/hg38/liftOver/
Chain File from hg19 (GRCh37) to hg17/hg18/hg38 and all other organisms: http://hgdownload.soe.ucsc.edu/goldenPath/hg19/liftOver/
Chain File from mm10 (GRCm38) to mm9 and all other organisms: http://hgdownload.soe.ucsc.edu/goldenPath/mm10/liftOver/
Ensembl chain files
Human to Human: ftp://ftp.ensembl.org/pub/assembly_mapping/homo_sapiens/
Mouse to Mouse: ftp://ftp.ensembl.org/pub/assembly_mapping/mus_musculus/
Other organisms: ftp://ftp.ensembl.org/pub/assembly_mapping/
User Input file
CrossMap supports the following file formats.
Output file
The format of output files depends on the input
Input_format |
Output_format |
|---|---|
BED |
BED (Genome coordinates will be updated) |
BAM |
BAM (Genome coordinates, header section, all SAM flags, insert size will be updated) |
CRAM |
BAM (require pysam >= 0.8.2) |
SAM |
SAM (Genome coordinates, header section, all SAM flags, insert size will be updated) |
Wiggle |
BigWig |
BigWig |
BigWig |
GFF |
GFF (Genome coordinates will be updated to the target assembly) |
GTF |
GTF (Genome coordinates will be updated to the target assembly) |
VCF |
VCF (header section, Genome coordinates, reference alleles will be updated) |
GVCF |
GVCF (header section, Genome coordinates, reference alleles will be updated) |
MAF |
MAF (Genome coordinates and reference alleles will be updated) |
Usage
Run CrossMap -h or CrossMap --help print help message
$ CrossMap -h
usage: CrossMap [-h] [-v]
{bed,bam,gff,wig,bigwig,vcf,gvcf,maf,region,viewchain} ...
CrossMap (v0.7.0) is a program to convert (liftover) genome coordinates
between different reference assemblies (e.g., from human GRCh37/hg19 to
GRCh38/hg38 or vice versa). Supported file formats: BAM, BED, BigWig, CRAM,
GFF, GTF, GVCF, MAF (mutation annotation format), SAM, Wiggle, and VCF.
positional arguments:
{bed,bam,gff,wig,bigwig,vcf,gvcf,maf,region,viewchain}
sub-command help
bed converts BED, bedGraph or other BED-like files. Only
genome coordinates (i.e., the first 3 columns) will be
updated. Regions mapped to multiple locations to the
new assembly will be split. Use the "region" command
to liftover large genomic regions. Use the "wig"
command if you need bedGraph/bigWig output.
bam converts BAM, CRAM, or SAM format file. Genome
coordinates, header section, all SAM flags, insert
size will be updated.
gff converts GFF or GTF format file. Genome coordinates
will be updated.
wig converts Wiggle or bedGraph format file. Genome
coordinates will be updated.
bigwig converts BigWig file. Genome coordinates will be
updated.
vcf converts VCF file. Genome coordinates, header section,
reference alleles will be updated.
gvcf converts GVCF file. Genome coordinates, header
section, reference alleles will be updated.
maf converts MAF (mutation annotation format) file. Genome
coordinates and reference alleles will be updated.
region converts big genomic regions (in BED format) such as
CNV blocks. Genome coordinates will be updated.
viewchain prints out the content of a chain file into a human
readable, block-to-block format.
options:
-h, --help show this help message and exit
-v, --version show program's version number and exit
https://crossmap.readthedocs.io/en/latest/
Convert BED format files
A BED (Browser Extensible Data) file is a tab-delimited text file describing genome regions or gene annotations. It consists of one line per feature, each containing 3-12 columns. CrossMap converts BED files with less than 12 columns to a different assembly by updating the chromosome and genome coordinates only; all other columns remain unchanged. Regions from the old assembly mapping to multiple locations to the new assembly will be split. For 12-columns BED files, all columns will be updated accordingly except the 4th column (name of bed line), 5th column (score value), and 9th column (RGB value describing the display color). 12-column BED files usually define multiple blocks (e.g., exons); if any of the exons fails to map to a new assembly, the whole BED line is skipped.
The input BED file can be plain text file, compressed file with extension of .gz, .Z, .z, .bz, .bz2 and .bzip2, or even a URL pointing to accessible remote files (http://, https:// and ftp://). Compressed remote files are not supported. The output is a BED format file with exactly the same number of columns as the original one.
Standard BED format has 12 columns, but CrossMap also supports BED-like formats:
BED3: The first three columns (“chrom”, “start”, “end”) of the BED format file.
BED6: The first six columns (“chrom”, “start”, “end”, “name”, “score”, “strand”) of the BED format file.
Other: Format has at least three columns (“chrom”, “start”, “end”) and no more than 12 columns. All other columns are arbitrary.
Note
For BED-like formats mentioned above, CrossMap only updates the “chrom”, “start”, “end”, and “strand” columns. All other columns will be kept AS-IS.
Lines starting with ‘#’, ‘browser’, ‘track’ will be skipped.
Lines less than three columns will be skipped.
The 2nd and 3rd columns must be integers.
The “+” strand is assumed if no strand information is found.
For standard BED format (12 columns). If any of the defined exon blocks cannot be uniquely mapped to target assembly, the whole entry will be skipped.
The “input_chain_file” and “input_bed_file” can be regular or compressed (.gz, .Z, .z, .bz, .bz2, .bzip2) file, local file or URL (http://, https://, ftp://) pointing to remote file.
If the output_file is not specified, results will be printed to screen (console). In this case, the original bed entries (including entries failed to convert) were also printed out.
If the input region cannot be consecutively mapped to the target assembly, it will be split.
The *.unmap file contains regions that cannot be unambiguously converted.
Typing CrossMap bed -h will print help message:
$ CrossMap bed -h
usage: CrossMap bed [-h] [--chromid {a,s,l}] [--unmap-file UNMAP_FILE]
input.chain input.bed [out_bed]
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or
compressed (.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.bed The input BED file. The first 3 columns must be
“chrom”, “start”, and “end”. The input BED file can be
plain text file, compressed file with extension of
.gz, .Z, .z, .bz, .bz2 and .bzip2, or even a URL
pointing to accessible remote files (http://, https://
and ftp://). Compressed remote files are not
supported.
out_bed Output BED file. if argument is missing, CrossMap will
write BED file to the STDOUT.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" =
"long style" (eg. "chr1", "chrX"); "s" = "short style"
(eg. "1", "X").
--unmap-file UNMAP_FILE
file to save unmapped entries. This will be ignored if
[out_bed] was not provided.
Example 1
run CrossMap bed with no output file.
Results were printed to screen.
$cat Test1.hg19.bed
chr1 65886334 66103176
chr3 112251353 112280810
chr5 54408798 54469005
chr7 107204401 107218968
$ CrossMap bed GRCh37_to_GRCh38.chain.gz Test1.hg19.bed
2024-01-12 08:36:35 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
chr1 65886334 66103176 -> chr1 65420651 65637493
chr3 112251353 112280810 -> chr3 112532506 112561963
chr5 54408798 54469005 -> chr5 55112970 55173177
chr7 107204401 107218968 -> chr7 107563956 107578523
Note
The first 3 columns are hg19-based coordinates. The last 3 columns are hg38-based coordinates.
Example 2
run CrossMap bed with output file specified.
Results (genomic coordinates after liftover) were saved to the file “output.hg38”
$ CrossMap bed GRCh37_to_GRCh38.chain.gz Test1.hg19.bed output.hg38
2024-01-12 08:35:56 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
$ cat output.hg38
chr1 65420651 65637493
chr3 112532506 112561963
chr5 55112970 55173177
chr7 107563956 107578523
Note
Genomic intervals failed to map will be saved to file “output.hg38.unmap”.
Example 3
Input regions will be split if they cannot map consecutively to the target assembly.
$ cat Test2.hg19.bed
chr20 21106623 21227258
chr22 30792929 30821291
$ CrossMap bed GRCh37_to_GRCh38.chain.gz Test2.hg19.bed
2024-01-12 08:53:10 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
chr20 21106623 21227258 (split.1:chr20:21106623:21144549:+) chr20 21125982 21163908
chr20 21106623 21227258 (split.2:chr20:21144549:21176014:+) chr20 21163909 21195374
chr20 21106623 21227258 (split.3:chr20:21176014:21186161:+) chr20 21195375 21205522
chr20 21106623 21227258 (split.4:chr20:21186161:21227258:+) chr20 21205523 21246620
chr22 30792929 30821291 (split.1:chr22:30792929:30819612:+) chr22 30396940 30423623
chr22 30792929 30821291 (split.2:chr22:30819615:30821291:+) chr22 30423627 30425303
Note
The first 3 columns are hg19-based coordinates. The last 3 columns are hg38-based coordinates.
Example 4
BedGraph format file can be converted using either CrossMap bed or CrossMap wig;
however, the output formats are different:
When using
CrossMap bedcommand to convert a bedGraph file, the output is a bedGraph file.When using
CrossMap wigcommand to convert a bedGraph file, the output is a bigWig file.
$ head -3 Test3.hg19.bgr
chrX 2705083 2705158 1.0
chrX 2813094 2813169 0.9
chrX 2813169 2814363 0.1
...
$ CrossMap bed hg19ToHg38.over.chain.gz Test3.hg19.bgr
2024-01-12 09:05:05 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz
chrX 2705083 2705158 1.0 -> chrX 2787042 2787117 1.0
chrX 2813094 2813169 0.9 -> chrX 2895053 2895128 0.9
chrX 2813169 2814363 0.1 -> chrX 2895128 2896322 0.1
...
$ CrossMap wig GRCh37_to_GRCh38.chain.gz Test3.hg19.bgr output.hg38
2024-01-12 09:09:52 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
2024-01-12 09:09:52 [INFO] Liftover wiggle file "Test3.hg19.bgr" to bedGraph file "output.hg38.bgr"
2024-01-12 09:09:52 [INFO] Merging overlapped entries in bedGraph file
2024-01-12 09:09:52 [INFO] Sorting bedGraph file: output.hg38.bgr
2024-01-12 09:09:52 [INFO] Writing header to "output.hg38.bw" ...
2024-01-12 09:09:52 [INFO] Writing entries to "output.hg38.bw" ...
Example 5
Use CrossMap region command to convert large genomic regions (such as CNV blocks) in BED format.
$ cat Test4.hg19.bed
chr2 239716679 243199373 # A large genomic interval of 3.48 Mb
If use the :code:`CrossMap bed`command to liftover, this interval will be split 74 times.
$ CrossMap bed GRCh37_to_GRCh38.chain.gz Test4.hg19.bed
2024-01-12 09:12:45 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
chr2 239716679 243199373 (split.1:chr2:239716679:239801978:+) chr2 238808038 238893337
chr2 239716679 243199373 (split.2:chr2:239831978:240205681:+) chr2 238910282 239283985
chr2 239716679 243199373 (split.3:chr2:240205681:240319336:+) chr2 239283986 239397641
... (split 74 times)
Use the CrossMap region command to liftover. By defualt r = 0.85.
$ CrossMap region GRCh37_to_GRCh38.chain.gz Test4.hg19.bed
2024-01-12 09:15:24 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
chr2 239716679 243199373 -> chr2 238808038 242183529 map_ratio=0.9
If we set r = 0.99, this region will fail.
$ CrossMap region GRCh37_to_GRCh38.chain.gz Test4.hg19.bed -r 0.99
2024-01-12 09:18:53 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
chr2 239716679 243199373 Fail map_ratio=0.9622
Convert BAM/CRAM/SAM format files
SAM (Sequence Alignment Map) format is a generic format for storing sequencing alignments, and BAM is the binary and compressed version of SAM (Li et al., 2009). CRAM was designed to be an efficient reference-based alternative to the SAM and BAM file formats. Most high-throughput sequencing (HTS) alignments were in SAM/BAM format and many HTS analysis tools work with SAM/BAM format. CrossMap updates chromosomes, genome coordinates, header sections, and all SAM flags accordingly. CrossMap’s version number is inserted into the header section, along with the names of the original BAM file and the chain file. For pair-end sequencing, insert size is also recalculated. The input BAM file should be sorted and indexed properly using Samtools (Li et al., 2009). The output format is determined by the input format, and the BAM output will be sorted and indexed automatically.
Typing CrossMap bam -h will print help message:
$ CrossMap bam -h
usage: CrossMap bam [-h] [-m INSERT_SIZE] [-s INSERT_SIZE_STDEV]
[-t INSERT_SIZE_FOLD] [-a] [--chromid {a,s,l}]
input.chain input.bam [out_bam]
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or
compressed (.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.bam Input BAM file (https://genome.ucsc.edu/FAQ/FAQformat.
html#format5.1).
out_bam Output BAM file. if argument is missing, CrossMap will
write BAM file to the STDOUT.
options:
-h, --help show this help message and exit
-m INSERT_SIZE, --mean INSERT_SIZE
Average insert size of pair-end sequencing (bp).
-s INSERT_SIZE_STDEV, --stdev INSERT_SIZE_STDEV
Stanadard deviation of insert size.
-t INSERT_SIZE_FOLD, --times INSERT_SIZE_FOLD
A mapped pair is considered as "proper pair" if both
ends mapped to different strand and the distance
between them is less then '-t' * stdev from the mean.
-a, --append-tags Add tag to each alignment in BAM file. Tags for pair-
end alignments include: QF = QC failed, NN = both
read1 and read2 unmapped, NU = read1 unmapped, read2
unique mapped, NM = read1 unmapped, multiple mapped,
UN = read1 uniquely mapped, read2 unmap, UU = both
read1 and read2 uniquely mapped, UM = read1 uniquely
mapped, read2 multiple mapped, MN = read1 multiple
mapped, read2 unmapped, MU = read1 multiple mapped,
read2 unique mapped, MM = both read1 and read2
multiple mapped. Tags for single-end alignments
include: QF = QC failed, SN = unmaped, SM = multiple
mapped, SU = uniquely mapped.
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" =
"long style" (eg. "chr1", "chrX"); "s" = "short style"
(eg. "1", "X").
Example
$ CrossMap bam -a GRCh37_to_GRCh38.chain.gz Test5.hg19.bam output.hg38
Add tags: True
Insert size = 200.000000
Insert size stdev = 30.000000
Number of stdev from the mean = 3.000000
Add tags to each alignment = True
2024-01-12 09:29:11 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
[E::idx_find_and_load] Could not retrieve index file for 'Test5.hg19.bam'
2024-01-12 09:29:11 [INFO] Liftover BAM file "Test5.hg19.bam" to "output.hg38.bam"
2024-01-12 09:29:15 [INFO] Done!
2024-01-12 09:29:15 [INFO] Sort "output.hg38.bam" and save as "output.hg38.sorted.bam"
2024-01-12 09:29:15 [INFO] Index "output.hg38.sorted.bam" ...
Total alignments:99914
QC failed: 0
Paired-end reads:
R1 unique, R2 unique (UU): 96035
R1 unique, R2 unmapp (UN): 3638
R1 unique, R2 multiple (UM): 0
R1 multiple, R2 multiple (MM): 0
R1 multiple, R2 unique (MU): 230
R1 multiple, R2 unmapped (MN): 11
R1 unmap, R2 unmap (NN): 0
R1 unmap, R2 unique (NU): 0
R1 unmap, R2 multiple (NM): 0
Optional tags:
- Q
QC. QC failed.
- N
Unmapped. Originally unmapped or originally mapped but failed to lift over to new assembly.
- M
Multiple mapped. Alignment can be lifted over to multiple places.
- U
Unique mapped. Alignment can be lifted over to only 1 place.
Tags for pair-end sequencing include:
QF = QC failed
NN = both read1 and read2 unmapped
NU = read1 unmapped, read2 unique mapped
NM = read1 unmapped, multiple mapped
UN = read1 uniquely mapped, read2 unmap
UU = both read1 and read2 uniquely mapped
UM = read1 uniquely mapped, read2 multiple mapped
MN = read1 multiple mapped, read2 unmapped
MU = read1 multiple mapped, read2 unique mapped
MM = both read1 and read2 multiple mapped
Tags for single-end sequencing include:
QF = QC failed
SN = unmaped
SM = multiple mapped
SU = uniquely mapped
Note
All alignments (mapped, partial mapped, unmapped, QC failed) will write to one file. Users can filter them by tags.
The header section will be updated to the target assembly.
Genome coordinates and all SAM flags in the alignment section will be updated to the target assembly.
If the input is a CRAM file, pysam version should >= 0.8.2
Optional fields in the alignment section will not update.
Convert Wiggle format files
Wiggle (WIG) format is useful in displaying continuous data such as GC content and the reads intensity of high-throughput sequencing data. BigWig is a self-indexed binary-format Wiggle file and has the advantage of supporting random access. Input wiggle data can be in variableStep (for data with irregular intervals) or fixedStep (for data with regular intervals). Regardless of the input, the output files are always in bedGraph format.
Typing CrossMap wig -h will print help message:
$ CrossMap wig -h
usage: CrossMap wig [-h] [--chromid {a,s,l}] input.chain input.wig out_wig
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or compressed
(.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.wig The input wiggle/bedGraph format file
(http://genome.ucsc.edu/goldenPath/help/wiggle.html).
Both "variableStep" and "fixedStep" wiggle lines are
supported. The input wiggle/bedGraph file can be plain
text file, compressed file with extension of .gz, .Z, .z,
.bz, .bz2 and .bzip2, or even a URL pointing to
accessible remote files (http://, https:// and ftp://).
Compressed remote files are not supported.
out_wig Output bedGraph file. Regardless of the input is wiggle
or bedGraph, the output file is always in bedGraph
format.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" = "long
style" (eg. "chr1", "chrX"); "s" = "short style" (eg.
"1", "X").
Note
To improve performance, this script calls GNU “sort” command internally. If the “sort” command is not callable, CrossMap will exit.
Convert BigWig format files
If an input file is in BigWig format, the output is BigWig format if UCSC’s ‘wigToBigWig’ executable can be found; otherwise, the output file will be in bedGraph format.
Typing CrossMap bigwig -h will print help message.:
$ CrossMap bigwig -h
usage: CrossMap bigwig [-h] [--chromid {a,s,l}] input.chain input.bw output.bw
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or compressed
(.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.bw The input bigWig format file
(https://genome.ucsc.edu/goldenPath/help/bigWig.html).
output.bw Output bigWig file.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" = "long
style" (eg. "chr1", "chrX"); "s" = "short style" (eg.
"1", "X").
Example
$ CrossMap bigwig GRCh37_to_GRCh38.chain.gz Test6.hg19.bw output.hg38
2024-01-12 09:37:32 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
2024-01-12 09:37:33 [INFO] Liftover bigwig file Test6.hg19.bw to bedGraph file output.hg38.bgr:
2024-01-12 09:37:33 [INFO] Merging overlapped entries in bedGraph file
2024-01-12 09:37:33 [INFO] Sorting bedGraph file: output.hg38.bgr
2024-01-12 09:37:33 [INFO] Writing header to "output.hg38.bw" ...
2024-01-12 09:37:33 [INFO] Writing entries to "output.hg38.bw" ...
Note
To improve performance, this script calls GNU “sort” command internally. If “sort” command does not exist, CrossMap will exit.
Convert GFF/GTF format files
GFF (General Feature Format) is another plain text file used to describe gene structure. GTF (Gene Transfer Format) is a refined version of GTF. The first eight fields are the same as GFF. Plain text, compressed plain text, and URLs pointing to remote files are all supported. Only chromosome and genome coordinates are updated. The format of the output is determined from the input.
Typing CrossMap gff -h will print help message:
$ CrossMap gff -h
usage: CrossMap gff [-h] [–chromid {a,s,l}] input.chain input.gff [out_gff]
- positional arguments:
- input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html) describes pairwise alignments between two genomes. The input chain file can be a plain text file or compressed (.gz, .Z, .z, .bz, .bz2, .bzip2) file.
- input.gff The input GFF (General Feature Format,
http://genome.ucsc.edu/FAQ/FAQformat.html#format3) or GTF (Gene Transfer Format, http://genome.ucsc.edu/FAQ/FAQformat.html#format4) file. The input GFF/GTF file can be plain text file, compressed file with extension of .gz, .Z, .z, .bz, .bz2 and .bzip2, or even a URL pointing to accessible remote files (http://, https:// and ftp://). Compressed remote files are not supported.
- out_gff Output GFF/GTF file. if argument is missing, CrossMap
will write GFF/GTF file to the STDOUT.
- options:
- -h, --help
show this help message and exit
- –chromid {a,s,l} The style of chromosome IDs. “a” = “as-is”; “l” = “long
style” (eg. “chr1”, “chrX”); “s” = “short style” (eg. “1”, “X”).
Example
$ CrossMap gff GRCh37_to_GRCh38.chain.gz Test7.hg19.gtf output.hg38
2024-01-12 09:43:53 [INFO] Read the chain file "GRCh37_to_GRCh38.chain.gz"
Note
Each feature (exon, intron, UTR, etc) is processed separately and independently, and we do NOT check if features originally belonging to the same gene were converted into the same gene.
If a user wants to lift over gene annotation files, use BED12 format.
If no output file was specified, the output will be printed to screen (console). In this case, items that failed to convert are also printed out.
Convert VCF format files
VCF (variant call format) is a flexible and extendable line-oriented text format developed by the 1000 Genome Project. It is useful for representing single nucleotide variants, indels, copy number variants, and structural variants. Chromosomes, coordinates, and reference alleles are updated to a new assembly, and all the other fields are not changed.
Typing CrossMap vcf -h will print help message:
$ CrossMap vcf -h
usage: CrossMap vcf [-h] [--chromid {a,s,l}] [--no-comp-alleles] [--compress]
input.chain input.vcf refgenome.fa out_vcf
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or compressed
(.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.vcf Input VCF (variant call format,
https://samtools.github.io/hts-specs/VCFv4.2.pdf). The
VCF file can be plain text file, compressed file with
extension of .gz, .Z, .z, .bz, .bz2 and .bzip2, or even a
URL pointing to accessible remote files (http://,
https:// and ftp://). Compressed remote files are not
supported.
refgenome.fa Chromosome sequences of target assembly in FASTA
(https://en.wikipedia.org/wiki/FASTA_format) format.
out_vcf Output VCF file.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" = "long
style" (eg. "chr1", "chrX"); "s" = "short style" (eg.
"1", "X").
--no-comp-alleles If set, CrossMap does NOT check if the reference allele
is different from the alternate allele.
--compress If set, compress the output VCF file by calling the
system "gzip".
Note
Genome coordinates and reference alleles will be updated to target assembly.
The reference genome is the genome sequences of target assembly.
If the reference genome sequence file (../database/genome/hg18.fa) was not indexed, CrossMap will automatically index it (only the first time you run CrossMap).
Output files: output_file and output_file.unmap.
In the output VCF file, whether the chromosome IDs contain “chr” or not depends on the format of the input VCF file.
Interpretation of Failed tags:
Fail(Multiple_hits) : This genomic location was mapped to two or more locations to the target assembly.
Fail(REF==ALT) : After liftover, the reference allele is the same as the alternative allele (i.e. this is NOT an SNP/variant after liftover). In version 0.5.2, this checking can be turned off by setting ‘–no-comp-alleles’.
Fail(Unmap) : Unable to map this location to the target assembly.
Fail(KeyError) : Unable to find the contig ID (or chromosome ID) from the reference genome sequence (of the target assembly).
Convert MAF format files
MAF (mutation annotation format) files are tab-delimited files that contain somatic and/or germline mutation annotations. Please do not confuse with the Multiple Alignment Format.
Typing CrossMap maf -h will print help message:
$ CrossMap maf -h
usage: CrossMap maf [-h] [--chromid {a,s,l}]
input.chain input.maf refgenome.fa build_name out_maf
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or compressed
(.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.maf Input MAF (https://docs.gdc.cancer.gov/Data/File_Formats/
MAF_Format/) format file. The MAF file can be plain text
file, compressed file with extension of .gz, .Z, .z, .bz,
.bz2 and .bzip2, or even a URL pointing to accessible
remote files (http://, https:// and ftp://). Compressed
remote files are not supported.
refgenome.fa Chromosome sequences of target assembly in FASTA
(https://en.wikipedia.org/wiki/FASTA_format) format.
build_name the name of the *target_assembly* (eg "GRCh38").
out_maf Output MAF file.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" = "long
style" (eg. "chr1", "chrX"); "s" = "short style" (eg.
"1", "X").
Convert GVCF format files
GVCF file format is described in here.
Typing CrossMap gvcf -h will print help message:
$ CrossMap gvcf -h
usage: CrossMap gvcf [-h] [--chromid {a,s,l}] [--no-comp-alleles] [--compress]
input.chain input.gvcf refgenome.fa out_gvcf
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or compressed
(.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.gvcf Input gVCF (genomic variant call format,
https://samtools.github.io/hts-specs/VCFv4.2.pdf). The
gVCF file can be plain text file, compressed file with
extension of .gz, .Z, .z, .bz, .bz2 and .bzip2, or even a
URL pointing to accessible remote files (http://,
https:// and ftp://). Compressed remote files are not
supported.
refgenome.fa Chromosome sequences of target assembly in FASTA
(https://en.wikipedia.org/wiki/FASTA_format) format.
out_gvcf Output gVCF file.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" = "long
style" (eg. "chr1", "chrX"); "s" = "short style" (eg.
"1", "X").
--no-comp-alleles If set, CrossMap does NOT check if the reference allele
is different from the alternate allele.
--compress If set, compress the output VCF file by calling the
system "gzip".
Convert large genomic regions
For large genomic regions such as CNV blocks, the CrossMap bed will split each large region into smaller blocks that are 100% matched to the target assembly.
CrossMap region will NOT split large regions, instead, it will calculate the map ratio (i.e. {bases mapped to target genome} / {total bases in query region}). If the
map ratio is larger than the threshold specified by -r, the coordinates will be converted to the target genome, otherwise, it fails.
Typing CrossMap region -h will print help message:
usage: CrossMap region [-h] [--chromid {a,s,l}] [-r MIN_MAP_RATIO]
input.chain input.bed [out_bed]
positional arguments:
input.chain Chain file
(https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The
input chain file can be a plain text file or
compressed (.gz, .Z, .z, .bz, .bz2, .bzip2) file.
input.bed The input BED file. The first 3 columns must be
“chrom”, “start”, and “end”. The input BED file can be
plain text file, compressed file with extension of
.gz, .Z, .z, .bz, .bz2 and .bzip2, or even a URL
pointing to accessible remote files (http://, https://
and ftp://). Compressed remote files are not
supported.
out_bed Output BED file. if argument is missing, CrossMap will
write BED file to the STDOUT.
options:
-h, --help show this help message and exit
--chromid {a,s,l} The style of chromosome IDs. "a" = "as-is"; "l" =
"long style" (eg. "chr1", "chrX"); "s" = "short style"
(eg. "1", "X").
-r MIN_MAP_RATIO, --ratio MIN_MAP_RATIO
Minimum ratio of bases that must remap.
Note
Input BED file should have at least 3 columns (chrom, start, end). Additional columns will be kept as is.
View chain file
Typing CrossMap viewchain -h will print help message:
usage: CrossMap viewchain [-h] input.chain
positional arguments:
input.chain Chain file (https://genome.ucsc.edu/goldenPath/help/chain.html)
describes pairwise alignments between two genomes. The input
chain file can be a plain text file or compressed (.gz, .Z, .z,
.bz, .bz2, .bzip2) file.
options:
-h, --help show this help message and exit
Example:
$ CrossMap viewchain GRCh37_to_GRCh38.chain.gz >chain.tab
$ head chain.tab
1 10000 177417 + 1 10000 177417 +
1 227417 267719 + 1 257666 297968 +
1 317719 471368 + 1 347968 501617 -
1 521368 1566075 + 1 585988 1630695 +
1 1566075 1569784 + 1 1630696 1634405 +
1 1569784 1570918 + 1 1634408 1635542 +
1 1570918 1570922 + 1 1635546 1635550 +
1 1570922 1574299 + 1 1635560 1638937 +
1 1574299 1583669 + 1 1638938 1648308 +
1 1583669 1583878 + 1 1648309 1648518 +
Compare to UCSC liftover tool
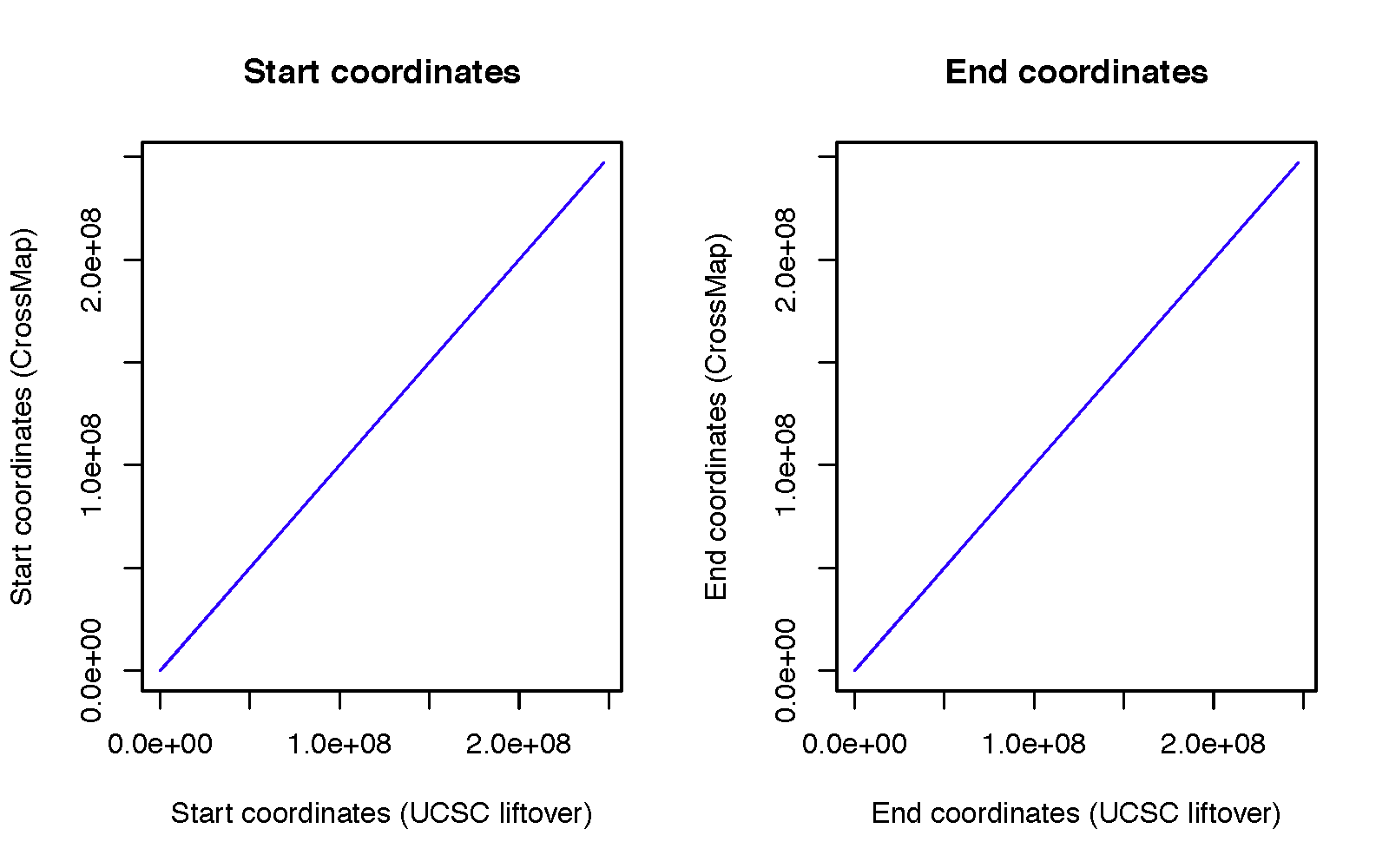
To assess the accuracy of CrossMap, we randomly generated 10,000 genome intervals (download from here) with the fixed interval size of 200 bp from hg19. Then we converted them into hg18 using CrossMap and UCSC liftover tool with default configurations. We compare CrossMap to UCSC liftover tool because it is the most widely used tool to convert genome coordinates.
CrossMap failed to convert 613 intervals, and the UCSC liftover tool failed to convert 614 intervals. All failed intervals are exactly the same except for one region (chr2 90542908 90543108). UCSC failed to convert it because this region needs to be split twice:
Original (hg19) |
Split (hg19) |
Target (hg18) |
|---|---|---|
chr2 90542908 90543108 - |
chr2 90542908 90542933 - |
chr2 89906445 89906470 - |
chr2 90542908 90543108 - |
chr2 90542933 90543001 - |
chr2 87414583 87414651 - |
chr2 90542908 90543108 - |
chr2 90543010 90543108 - |
chr2 87414276 87414374 - |
For genome intervals that were successfully converted to hg18, the start and end coordinates are exactly the same between UCSC conversion and CrossMap conversion.
Citation
LICENSE
CrossMap is distributed under GNU General Public License
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301 USA
Contact
Wang.Liguo AT mayo.edu